Home> News
New technology helps improve RNA-DNA interactions analysis
Updated: 2017-10-25
Zhong Sheng, a professor with the University of California San Diego (UCSD), has developed MARGI (Mapping RNA Genome Interactions), a new tool to reveal RNA-DNA interactions. The technology enables a single experiment to completely record all RNA molecules interacting with a segment of DNA and the locations of all these interactions. The study will help researchers quickly decipher the gene sequences’ functions based on that information. The findings -- “Systematic mapping of RNA-chromatin interactions in vivo” -- were recently published in Current Biology, a subsidiary journal of Cell.
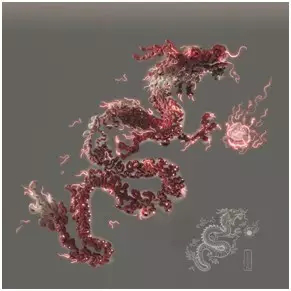
An illustration of RNA-DNA interactions [Victor O. Leshyk/From reference 2]
Victor O. Leshyk used a dragon-like image to demonstrate the RNA-DNA interactions. A 3-D structure of tightly coiled DNA is depicted as the body of a dragon in Chinese myth. Interacting RNAs are depicted as hairs, whiskers and claws, which are essential for the dragon to function.
RNA molecules can attach to particular DNA sequences and help control the amount of proteins produced by these particular genes in specific cells and at specific times. Researchers can identify the new functions of the genome’s code by determining which genes are involved in generating regulatory RNA.
The corresponding author of the research, Zhong Sheng, said most of the human genome sequences are known, but we still don’t know their functions and significance. In order to better understand those unknown functions, the researchers developed this new tool that provides information about all RNA molecules interacting with DNA and the interacted sequences.
The existing methods for RNA-DNA interactions research can only analyze the interaction between a RNA molecule and DNA in a single experiment. It is extremely tedious when the research involves hundreds of RNA molecules.
Using the traditional methods, the study would take years; with the help of MARGI, a single experiment can analyze all the RNA-DNA interactions in one or two weeks, according to Tri Nguyen, a bioengineering Ph.D. student at UCSD and a co-first author of the study.
The MARGI technology starts out with a mixture containing RNA and DNA that has been cut into short pieces. In this mixture, a subset of RNA molecules interacts with particular DNA pieces. A specially designed linker is then added to connect the interacting RNA-DNA pairs. Linked RNA-DNA pairs are selectively fished out, then converted into chimeric sequences that can all be read at once using high-throughput sequencing.
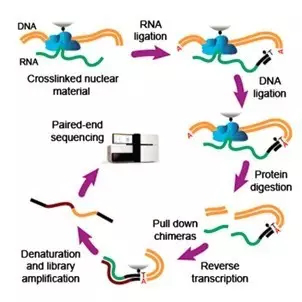
The MARGI operation diagram [From reference 1]
The researchers tested the method's accuracy by seeing if it produced false positive results. First, they mixed RNA and DNA from both fruit fly and human cells, creating both "true" RNA-DNA pairs, meaning they're either fully human or fully fruit fly, and "false" RNA-DNA pairs, meaning they're half human and half fruit fly -- these are the ones that shouldn't be detected. The team then screened the entire mixture using MARGI. The method detected a large set of true RNA-DNA interactions, and approximately 2 percent of MARGI-identified interactions were false.
It’s not the first time for Zhong to combine a traditional Chinese art form with achievements in scientific research on the cover of international magazines.
Early in 2014, he used the Eight Diagrams, or Bagua, a philosophical and ritual tradition of Chinese origin which embodies the idea of living in harmony, to describe the interactions between two embryonic mouse cells when he published his findings in Genome Research.

An illustration of the interactions between two embryonic mouse cells [From reference 3]
Zhong graducated from Peking University with a double major in Mathematics and Economics in 2001 and obtained a Ph.D. in Biostatistics at Harvard University in 2004. He worked at the BioX of Stanford University as a visiting scholar and in 2005 was employed by the University of Illinois Urbana-Champaign before moving to UCSD. He is also a winner of an NIH Pioneer Award, an NSF CAREER Award and an Alfred Sloan Fellowship. He received an Award for Excellence in Consulting from the University of Illinois Engineering Council for two consecutive years.

Professor Zhong [ucsd.edu]
We are expecting more scientists of Chinese origin to make great contributions in their research and promote traditional Chinese culture and arts.
References:
1. Sridhar B,Rivas-Astroza M, Nguyen TC, et al. (2017) Systematic mappingof RNA-chromatin interactions in vivo.Curr Biol 27: 602-709
2. Decoding the genome's cryptic language. (2017) https://www.eurekalert.org/pub_releases/2017-02/uoc--dtg022417.php
3. Biase FH, Cao X, Zhong S.(2014) Cell fate inclination within 2-cell and 4-cell mouse embryos revealed bysingle-cell RNA sequencing. Genome Res 24: 1787-1796
